Getting Started
Important
It is strongly recommended to use curifactory from within a git repo to
support experiment reproducibility and provenance (every run will record the
current git commit hash.) The curifactory init command will prompt you
to run git init if the .git folder is not detected. Any experiment
runs executed outside of a git repo will carry an associated warning in the
output log and report.
Basic components
This section follows the 0_BasicComponents notebook. We first introduce the four basic components of curifactory and show how to use them from a within a live context. (A live context is either a notebook, REPL, or non-experiment script.)
These components are:
the artifact manager
parameters
records
stages
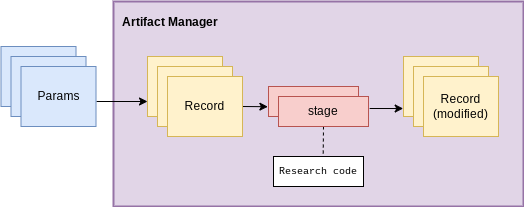
Artifact manager
First up is the artifact manager. ArtifactManager is a class
representing a session that tracks metadata and generated objects from
experiment code.
When an artifact manager is instantiated, it creates a new “experiment run”, which will have an associated log file and can generate a run-specific report.
from curifactory import ArtifactManager
manager = ArtifactManager("notebook_example_0") # we pass in a name to associate with this run.

The artifact manager contains and keeps track of the overall session for the experiment stuff.
Parameters
The next component is parameters. Parameter classes are a way to define how research code can be configured, e.g. how many layers to include in a neural network. By defining and initializing these directly in python, we have the ability to dynamically create experiment parameter configurations, compose them, etc.
Curifactory comes with an ExperimentParameters base class that all
custom parameter classes should inherit from. ExperimentParameters
includes a name parameter, allowing you to provide a label for the
parameter sets.
Note
For best ergonomics, we recommend defining an parameter class with the python
@dataclass decorator. This makes it easy to define defaults and
quickly view your parameter definitions simply as a collection of
attributes.
Parameter sets can then be initialized, passed around, and used within your research code, making it easy to organize and keep track of hyperparameters.
from dataclasses import dataclass
from curifactory import ExperimentParameters
@dataclass
class MyParams(ExperimentParameters):
some_scalar_multiplier: float = 1.0
# define a couple argument sets
default_params = MyParams(name="default")
doubled_params = MyParams(name="doubled", some_scalar_multiplier=2.0)
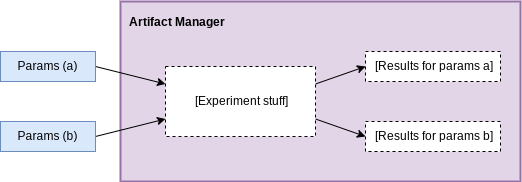
Creating and passing different parameter sets through the experiment stuff will give us different sets of results in the manager we can compare.
Records
A record is how curifactory keeps track of state in an experiment
run. “State” includes the data, objects, and results associated with a specific
parameter set, e.g. a trained model that came from using a particular
MyParams instance. The Record class is initialized with the
current manager as well as the argument set to use. Records
have a state dictionary, which holds intermediate data and objects
as research code is evaluated.
from curifactory import Record
r0 = Record(manager, default_params)
r1 = Record(manager, doubled_params)
print(r0.state, r0.params)
#> {} MyParams(name='default', hash=None, overwrite=False, some_scalar_multiplier=1.0)
print(r1.state, r1.params)
#> {} MyParams(name='doubled', hash=None, overwrite=False, some_scalar_multiplier=2.0)
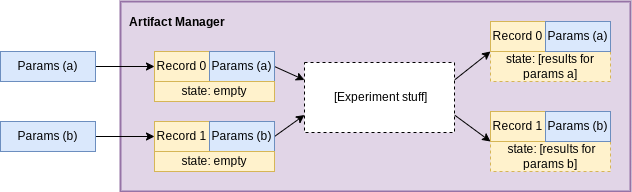
The state for each record is what’s actually storing the results from the experiment stuff for each given parameter set.
Stages
A stage represents a small, well-defined abstraction around portions
of research code which process some set of inputs and create a set of
outputs. A stage acts on a given record by taking the requested
inputs from that record’s state, evaluating some code, and returning values
that are then stored in the record’s state. This is implemented with a
@stage decorator which takes an array of string input names and an
array of string output names. Functions with the @stage decorator
must accept a record as the first argument.
Inside the stage, the record object that is passed in can be used to get
the current parameter set and parameterize the computation,
via the record.params attribute.
In the example below, we’ve defined a very simple stage that will store a number in the record’s state under the “initial_value” key.
Running a stage works by calling the function and passing it the record. The record itself is changed in-place, but it is also directly returned from the stage call. This allows functionally chaining stages, which we demonstate later on.
from curifactory import stage
@stage(inputs=None, outputs=["initial_value"])
def get_initial_value(record):
my_value = 5
return my_value * record.params.some_scalar_multiplier
r0 = get_initial_value(r0)
r1 = get_initial_value(r1)
After running both records through our stage, printing the states shows
the returned initial_value data.
print(r0.state, r1.state)
#> {'initial_value': 5.0} {'initial_value': 10.0}

Stages are the “experiment stuff”. A stage uses and modifies the state of a passed record. This is where the experiment code actually runs, taking any necessary inputs from the passed record state and storing any returned outputs back into it.
Specifying inputs on the stage decorator tells curifactory to search for those keys in the state of the passed record. Those values are then injected into the record call as kwargs. Note that the argument names in the function definition must match the string values of the inputs array.
"initial_value" was added to the state from the
get_initial_value stage, so we implement a stage below that expects
that piece of data and computes a new value based on it.
@stage(inputs=["initial_value"], outputs=["final_value"])
def multiply_again(record, initial_value):
return initial_value * record.params.some_scalar_multiplier
r1 = multiply_again(r1)
print(r1.state)
#> {'initial_value': 10.0, 'final_value': 20.0}
As mentioned before, since a stage accepts and returns a record, stages can be functionally chained together:
r2 = Record(manager, MyParams(name="uber-double", some_scalar_multiplier=4.0))
r2 = multiply_again(get_initial_value(r2))
print(r2.state, r2.params)
#> {'initial_value': 20.0, 'final_value': 80.0} MyParams(name='uber-double', hash=None, overwrite=False, some_scalar_multiplier=4.0)
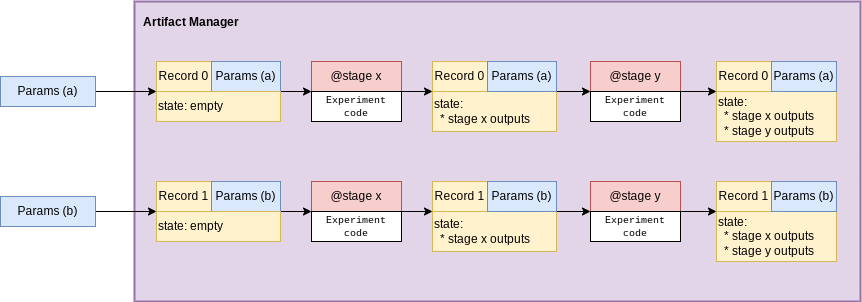
Records can be “pipelined” through sequences of stages to create a full experiment composed of larger abstract steps.
Records and stages represent linear chains of compute steps, but
in many cases it’s important to compare results and data across multiple pieces
of an experiment run (e.g. comparing the scores of an SVM with the
scores of a logistic regression algorithm.) @aggregate decorated
functions are a special kind of stage that additionally take a collection of
records to compute over. Aggregate stages still produce outputs and both take
and return a single record associated with it, meaning additional regular
stages can be chained after an aggregate stage.
We specify inputs to an @aggregate decorator the same way we do with
@stage, with a list of string names of artifacts from record state, and the
function definition still needs to have correspondingly named arguments for
those inputs. However, since an aggregate takes multiple records, these input
arguments are populated with dictionaries where each key is a record that has
the requested input in its state, and the value is that object in the state. Any
records that don’t have the requested input will throw a warning, and will be
absent from that dictionary.
@aggregate decorated stages must take the single record as the first parameter
(like a normal stage,) and the collection of records to compute over as the
second, followed by the arguments for any specified inputs.
In the example below, we iterate through the records to create a
dictionary of all associated final_value entries from each record’s
state, and then determine the maximum.
from curifactory import aggregate
@aggregate(inputs=["final_value"], outputs=["all_final_values", "maximum_value"])
def find_maximum_final_value(record, records, final_value: dict[Record, float]):
all_vals = {}
for r, value in final_value.items():
all_vals[r.params.name] = value
maximum = max(all_vals.values())
return all_vals, maximum
Sometimes an aggregate doesn’t really need its own parameter set, e.g. if it’s
simply comparing results from other records. In these cases, records can be initialized with None passed as the
parameter set. In the cell below, we manually pass our previous records into
the stage, but note that if we pass None for records (the default) it will take all existing records in the manager.
final_record = Record(manager, None)
final_record = find_maximum_final_value(final_record, [r0, r1, r2])
print(final_record.state)
#> {'all_final_values': {'doubled': 20.0, 'uber-double': 80.0}, 'maximum_value': 80.0}
Note that we ran our aggregate stage on three records, the first one of which
(r0) did not have a final_value artifact in state. While r0 will still be
passed in the records list that the aggregate stage has access to, the
final_value dictionary only had entries for r1 and r2, so the output
artifact all_final_values only lists those two parameter sets.

To recap, the artifact manager keeps track of the overall session for a run, the “experiment run container”. Parameter sets are created with different hyperparameters to test a hypothesis or vary the experiment. Records track state changes and intermediate data associated with a parameter set throughout the experiment. Stages are what modify record state, they apply research code to the passed records based on their associated parameters, and the results for each stage are stored back into the record’s now modified state.

Caching and reporting
This section follows the 1_CachingAndReporting notebook. Here we demonstrate some features the previously discussed components enable. Two major abilities are easily caching objects (to short circuit computation of already-computed values) and quickly adding graphs and other “reportables” to a jupyter display or a generated HTML experiment run report.
First we create an artifact manager, an args class, and some arg sets like in the previous example:
from dataclasses import dataclass
import curifactory as cf
manager = cf.ArtifactManager("notebook_example_1")
@dataclass
class Params(cf.ExperimentParameters):
my_parameter: int = 1
default_params = Params(name="default")
doubled_params = Params(name="doubled", my_parameter=2)
Caching
Caching is done at each stage by listing a
curifactory.Cacheable subclass for each output. After the stage
runs, each cacher will save the returned object in the data cache path.
The cached filename includes the name of the experiment (the string passed
to ArtifactManager, “notebook_example_1” in this case), the hash
string of the parameters (see the Hashing Mechanics page for more information on
how this gets calculated), the name of the stage doing the caching, and
the name of the output itself.
On any subsequent run of that stage, the cachers all check to see if their file has already been created, and if it has, they directly load the object from file and return it rather than running the stage code.
The @stage decorator has a cachers parameter which should be
given a list of the cachers to use for the associated
outputs list. Curifactory comes with a set of default cachers you can
use, including JsonCacher, PandasCSVCacher,
PandasJsonCacher, and PickleCacher.
In the example below, we define a “long-running compute” stage, to demonstrate cachers short-circuiting computation:
from time import sleep
from curifactory.caching import JsonCacher
@cf.stage(inputs=None, outputs=["long-compute-data"], cachers=[JsonCacher])
def long_compute_step(record):
some_data = {
"my_value": record.params.my_parameter,
"magic_value": 42
}
sleep(5) # making dictionaries is hard work
return some_data
We run a record through our long running stage, and as expected it takes 5 seconds:
%%time
r0 = cf.Record(manager, default_params)
r0 = long_compute_step(r0)
CPU times: total: 0 ns
Wall time: 5 s
Inspecting our cache path now, there’s a new json entry for our output, which we can load up and see is the output from our stage:
import json
print(os.listdir("data/cache"))
print()
with open(f"data/cache/{os.listdir('data/cache')[0]}", 'r') as infile:
print(json.load(infile))
['notebook_example_1_c504fab1c3ccad16d1e3ef540001172c_long_compute_step_long-compute-data.json']
{'my_value': 1, 'magic_value': 42}
If we run the stage again with a record using the same parameter set as the previous time, it finds the correct cached output and returns before running the stage code:
%%time
r1 = cf.Record(manager, default_params)
r1 = long_compute_step(r1)
CPU times: total: 0 ns
Wall time: 0 ns
Using a different parameter set results in a different cache path, so computations with different parameters won’t conflict:
r2 = cf.Record(manager, doubled_params)
r2 = long_compute_step(r2)
os.listdir("data/cache")
['notebook_example_1_2c48da4b242c95c4eafac7e88872d319_long_compute_step_long-compute-data.json',
'notebook_example_1_c504fab1c3ccad16d1e3ef540001172c_long_compute_step_long-compute-data.json']
Lazy loading
One potential pitfall with caching is that it will always load
the object into memory, even if that object is never used. Projects with
very large data objects can run into memory problems as a result.
Curifactory includes a Lazy class that can wrap around a stage
output string name - when it is first computed, the cacher saves it and
the object is removed from memory (replaced in the record state with a
Lazy instance.) When the lazy object is accessed, it will reload the
object into memory from cache at that point.
This means that in a sequence of stages where all values are cached, earlier stage outputs may never need to load into memory at all.
from curifactory.caching import Lazy
import sys
@cf.stage(inputs=None, outputs=[Lazy("very-large-object")], cachers=[JsonCacher])
def make_mega_big_object(record):
mega_big = [1]*1024*1024
print(sys.getsizeof(mega_big))
return mega_big
r3 = cf.Record(manager, default_params)
r3 = make_mega_big_object(r3)
r3.state.resolve = False
print(type(r3.state['very-large-object']))
print(sys.getsizeof(r3.state['very-large-object']))
<class 'curifactory.caching.Lazy'>
48
Note that Record.state is actually a custom subclass of dict,
and by default it will automatically resolve lazy objects any time they’re
accessed on the state. the above cell turns this functionality off (with
state.resolve = False) to show that what’s actually in memory before
a resolved access is just the lazy object, which is significantly
smaller.
When the record’s state resolve is at it’s default value of True:
r3.state.resolve = True
print(type(r3.state['very-large-object']))
print(sys.getsizeof(r3.state['very-large-object']))
<class 'list'>
8697456
Reporting
A major part of experiments for debugging, understanding, and
publishing them is the ability to present results and pretty graphs! This can be a
challenge to keep organized, as one tries to manage folders for
matplotlib graph images, result tables, and so on. Curifactory provides
shortcuts to easily create Reportable items from inside stages,
which the artifact manager can then display inside an experiment run report in its own uniquely named
run folder, which contains all of the information about the run, all of
the created reportables, and a map of the stages that were run. Many of
these report components can be rendered inside a notebook as well.
Every record has a report function that takes a Reportable
subclass. Curifactory includes multiple default reporters, such as
DFReporter, FigureReporter, HTMLReporter, JsonReporter,
and LinePlotReporter.
from curifactory.reporting import LinePlotReporter
@cf.stage(inputs=None, outputs=["line_history"])
def make_pretty_graphs(record):
multiplier = record.params.my_parameter
# here we just make a bunch of example arrays of data to plot
line_0 = [1 * multiplier, 2 * multiplier, 3 * multiplier]
line_1 = [3 * multiplier, 2 * multiplier, 1 * multiplier]
line_2 = [4, 0, 3]
# a LinePlotReporter makes a nicely formatted matplotlib graph
record.report(LinePlotReporter(line_0, name="single_line_plot"))
record.report(LinePlotReporter(
y={
"ascending": line_0,
"descending": line_1,
"static": line_2
},
name="multi_line_plot"
))
return [line_0, line_1, line_2]
r4 = cf.Record(manager, default_params)
r5 = cf.Record(manager, doubled_params)
r4 = make_pretty_graphs(r4)
r5 = make_pretty_graphs(r5)
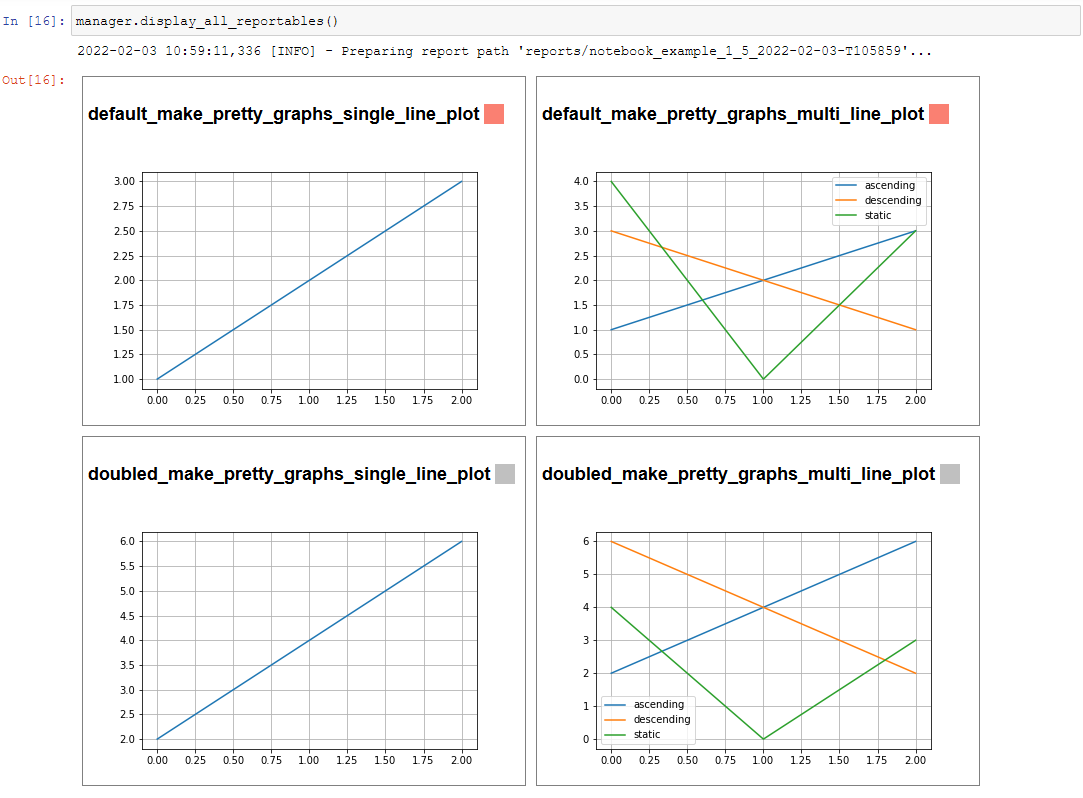
The example stage above adds a couple simple line plots to any record that is run through it.
When inside of a jupyter notebook or jupyter lab, the manager includes several display functions that allow you to render portions of the report directly in the notebook.
A few of these are:
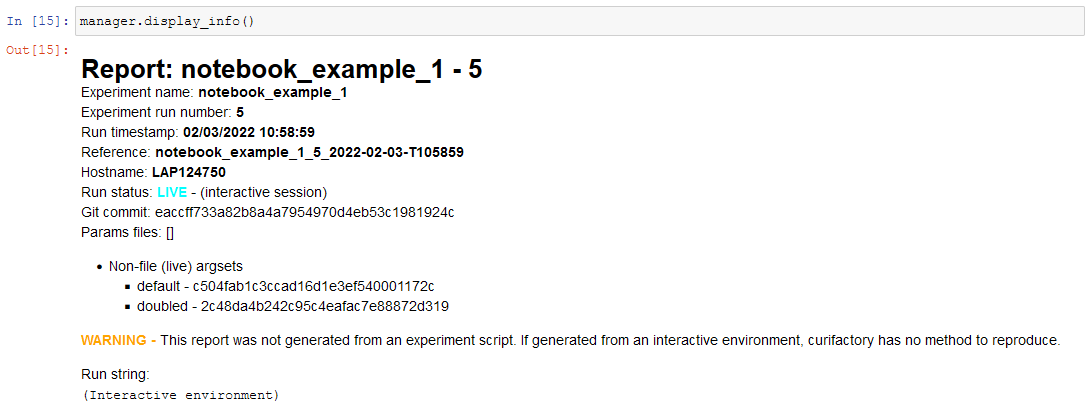
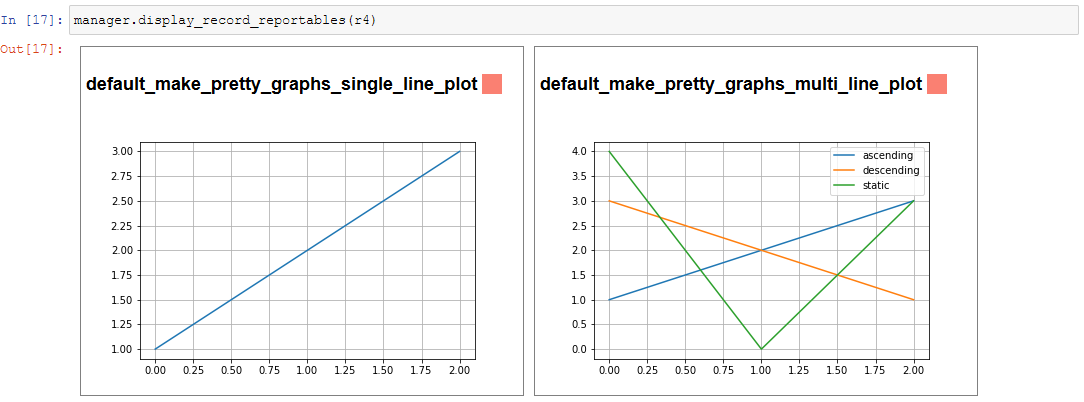
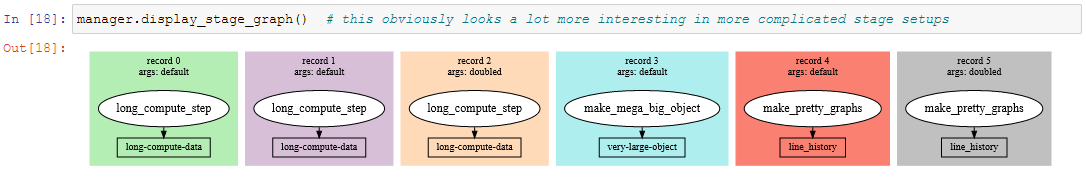
display_info()- renders the top block of the report, containing metadata about the rundisplay_all_reportables()- renders all reportables in the managerdisplay_record_reportables(record)- renders only the reportables associated with the passed recorddisplay_stage_graph()- renders a diagram of all the records, state objects, and stages. Note that graphviz must be installed for these to generate correctly.
Finally, a full HTML report can be produced with the
generate_report() function. This will create a run-specific folder
to contain the report and all rendered reportables, inside the reports
path. Additionally, every time a report is generated, an overall project
report index is put directly in the reports path, which lists and links
to all of the individual reports.
manager.generate_report()
2022-02-03 10:59:11,821 [INFO] - Generating report...
2022-02-03 10:59:11,821 [INFO] - Preparing report path 'reports/_latest'...
2022-02-03 10:59:12,360 [INFO] - Preparing report path 'reports/notebook_example_1_5_2022-02-03-T105859'...
2022-02-03 10:59:12,945 [INFO] - Updating report index...
2022-02-03 10:59:12,945 [INFO] - 2 labeled reports found
2022-02-03 10:59:12,945 [INFO] - 0 informal runs found
os.listdir("reports")
['index.html',
'notebook_example_1_4_2022-02-03-T092555',
'notebook_example_1_5_2022-02-03-T105859',
'style.css',
'_latest']
For more information on reports, see the Reports section.
Experiment organization
While the above sections demonstrate how to use curifactory in
notebooks or a python shell, most of the power of curifactory comes from its ability to help
organize experiment scripts and conduct more formal experiment runs with the
included experiment CLI tool.
Basic mechanics
An official experiment in curifactory fundamentally relies on two functions:
A
run()function that defines the experiment code to execute. This is where the code for creating records and running them through stages as shown in the previous sections would go.One (or more)
get_params()function(s) that return a list of arguments to apply to an experiment run.
When these two functions are in place, curifactory takes the list of arguments
computed from get_params() and passes it into the run() function
along with a fully initialized ArtifactManager.
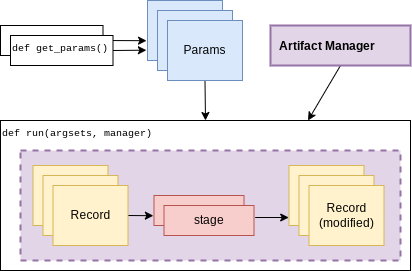
These mechanics provide a methodical way of creating curifactory-based runnables for a research project, and are what allows curifactory to inject its features into each experiment run (e.g. automatic logging, reporting, and a single CLI interface for interacting with the experiment runs.)
The experiment script file
So where do these functions go?
At its core, curifactory’s usage is based around “experiment scripts”, which are
python files in the experiment module path (part of curifactory’s configuration
established with the curifactory init command, by default this is a folder in
the project root experiments/, see Configuration and directory structure).
Experiment scripts must implement the aforementioned run() function, which takes
a list of ExperimentParameters subclass instances and an ArtifactManager:
from typing import List
import curifactory as cf
def run(param_sets: List[cf.ExperimentParameters], manager: cf.ArtifactManager):
# 1. make records
# 2. run stages
# 3. ???
# 4. PROFIT!
Stage implementations can go anywhere in your codebase, but for our simple example (or perhaps for stages that are only ever relevant to a specific experiment) we can include the stages directly in our experiment file. An example experiment setup with some stages might look like:
import curifactory as cf
from curifactory.caching import PickleCacher, JsonCacher
@cf.stage(inputs=None, outputs=["training_data", "testing_data"], cachers=[PickleCacher]*2):
def load_data(record):
# ...
@cf.stage(inputs=["training_data"], outputs=["model"], cachers=[PickleCacher])
def train_model(record, training_data):
# ...
@cf.aggregate(outputs=["scores"], cachers=[JsonCacher])
def test_models(record, records):
# ...
def run(param_sets, manager):
for param_set in param_sets:
record = cf.Record(manager, param_set)
train_model(load_data(record))
test_models(cf.Record(manager, None))
We can add in the parameters class that the stages need, and then make a basic
get_params() function to give us some parameter sets to compare a logistic
regression model versus a random forest classifier.
from dataclasses import dataclass
from sklearn.base import ClassifierMixin
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
import curifactory as cf
from curifactory.caching import PickleCacher, JsonCacher
@dataclass
class Params(cf.ExperimentParameters):
balanced: bool = False
"""Whether class weights should be balanced or not."""
n: int = 100
"""The number of trees for a random forest."""
seed: int = 42
"""The random state seed for data splitting and model training."""
model_type: ClassifierMixin = LogisticRegression
"""The sklearn model to use."""
def get_params():
return [
Params(name="simple_lr", balanced=True, model_type=LogisticRegression, seed=1),
Params(name="simple_rf", model_type=RandomForestClassifier, seed=1),
]
# ...
# stages
# ...
def run(param_sets, manager):
for param_set in param_sets:
record = cf.Record(manager, param_set)
train_model(load_data(record))
test_models(cf.Record(manager, None))
The fully implemented example shown here can be found at Example Experiment, as well as in the curifactory codebase under
examples/minimal/experiments/iris.py.
The experiment CLI
Curifactory’s main tool is the experiment CLI. This tool provides an
easy way to interact with your experiment code and control different
aspects of the experiment runs.
The experiment ls subcommand will list out your experiment
directory and parameter files (discussed later) and check for any basic errors.
Running this in our project’s root directory (assuming the above example is in
the experiment’s directory and named iris.py) we should get:
$ experiment ls
EXPERIMENTS:
iris
PARAMS:
iris
Essentially, this gives us the names of files that respectively have a
run() function (under EXPERIMENTS) and files that have a
get_params() function (under PARAMS).
We can execute an experiment by running experiment [EXPERIMENT_NAME] -p
[PARAMS_NAME]
In our case, the iris.py file has both functions (and appears under both
sections for experiment ls), so we can execute it with: (note you do not
include the file extension, you’re providing it as a module name.)
experiment iris -p iris
Curifactory has a shortcut for when an experiment file also has its own
get_params() function, you can equivalently just run:
experiment iris
which curifactory will internally expand to experiment [EXPERIMENT_NAME] -p
[EXPERIMENT_NAME]
The CLI has a large collection of flags to alter and control an experiment run, see the CLI Guide for more information.
Parameter files
As shown so far, for simplicity, experiments can have their own
get_params() function. However, frequently you may want to define multiple
different collections of parameter sets and selectively include them in an experiment, or
define sets that could be shared across multiple experiment scripts. One option
is to simply explicitly refer to a previous experiment file that has the
get_params() you want, e.g. with experiment iris_updated -p iris.
The other option is to create separate parameter files, or distinct files in the
params module path, (by default params/ in the project root.) These
files should each contain a get_params() function.
Importantly, you can specify multiple -p flags to the experiment CLI, like:
experiment iris -p params_file1 -p params_file2 -p iris
This will call the get_params() of every requested parameters file (and/or experiment
file) and combine all returned parameter sets into a single list that gets passed
into the experiment’s run().
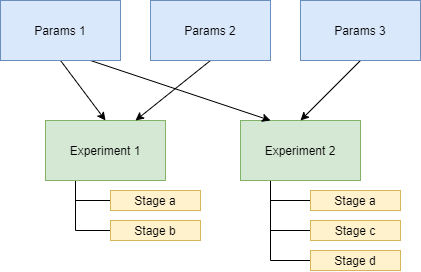
Creating distinct parameters files allows for structuring shared parameter sets to apply across experiments
Organization of growing projects
The structure and organization of experiments, parameters, and stages are relatively flexible, intended to allow the researcher to organize these however best to suit the needs of the project they’re working on.
The only organization constraints include:
Formal experiment scripts with
run()functions must go in the experiment module folder. (experiments/by default.)Formal argument set creation with
get_params()functions must either go in an experiment file or in python files in the params module folder. (params/by default.)
Lacking any additional constraints, some ideas for use include:
Stages can either be directly in the research codebase wrapping research functions, or they can remain separated and just make the appropriate calls into your codebase, parameterized with relevant args.
Keeping all parameters commonly used by all experiments can be kept in
params/__init__.py. If there are stages/params relevant only to a single experiment, these can be kept in the experiment file, and extracted out later if they become more generally useful. (Aparams.__init__.Paramsclass could be further subclassed in an experiment file to get the benefit of both.)Common stage sequences can be simplified and extracted into a helper file, e.g. by defining a function that takes a record, calls the stages, and returns the record from the last one:
def some_non_stage_helper_function(record):
return my_last_stage(my_middle_stage(my_first_stage(record)))
See the Tips and tricks for more ideas.
Next steps
Look through:
Components for a more in-depth understanding of the components and how they interact with each other.
Parameter files and parameter sets for fancier things you can do with parameters.
Cache for how to make custom cachers.
Reports to get an idea for how reports work and how to use them, plus how to make custom reportables.
CLI Guide for how to use the
experimentCLI program and what you can do with it.Tips and tricks for various “patterns” of use for Curifactory.