Tips and tricks
Minimum code to run stages
Directly/manually running stages can be useful for a variety of reasons, whether for debugging them, using the same configuration values as from an experiment, or as a basis for exploration in a jupyter notebook.
The smallest set of components needed to do this is an Params instance, an
ArtifactManager, and a Record:
# required components
from curifactory import ArtifactManager, Record
from params import Params # a custom dataclass extending cf.ExperimentParameters
# import whatever stages you need to run
from stages.data import get_data, clean_data
# initialize instances of the required components
params = Params() # create/initialize the params. Note that you could also directly
# load and manually call the get_params() of a parameter file
mngr = ArtifactManager("some_cache_ref_name") # the caching prefix name (normally
# the experiment name)
record = Record(mngr, params)
# run desired stages
clean_data(get_data(record))
# get the return from the last run stage (you could also get it from record.state)
print(record.output)
Note that calling any stage, e.g. get_data(record) returns the modified record that
was passed in, and since calling a stage always only takes exactly one parameter, (the record
to use) you can directly chain the stages together with clean_data(get_data(record))
Common dataset for multiple different argsets
For some experiments, you may have a single dataset processing procedure that all other parameter sets share. By default, caching only applies to a single parameter set, so the data would have to be recomputed for every parameter set, if you defined your entire experiment in one procedure. However, it’s possible to write your experiment to “share” the state of a data processing record with the remainder of your argsets.
def run(param_sets: List[Params], mngr: ArtifactManager):
# define the procedures
common_data_proc = Procedure([get_data, clean_data], mngr) # the resulting state of this procedure
# should be the same for all our
# parameter sets, so we only want to run
# once it
model_proc = Procedure([train_model, test_model], mngr) # this procedure is where we expect our
# argsets to differ, so run it per param set
# run our common procedure once (assuming all of the relevant parameter sets are _actually_ the
# same, we can just pull from the first one)
data_record = common_data_proc.run(param_sets[0])
# run each argset through model procedure
for param_set in param_sets:
record = data_record.make_copy(param_set) # duplicate the data record's state but with new param sets
model_proc.run(param_set, record) # we can manually specify the record to use in a procedure if
# we already have one
The above will result in an experiment graph that looks something like the following, assuming two argsets:
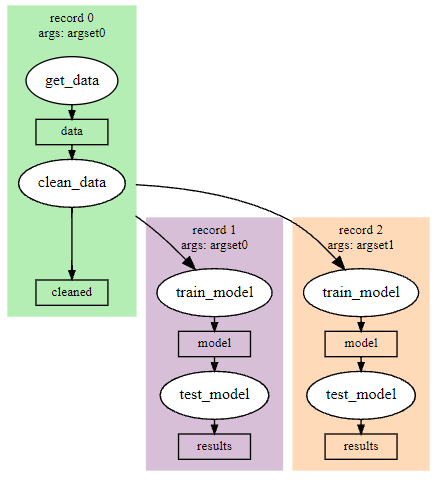
The make_copy() function on Record instances creates a new record with a deepcopy of the
state attribute, meaning you can define procedures that only have later stages, and pass them
the copied records.
This can be extended beyond just common datasets - any baseline procedure that will not change state across different parameter sets in a given experiment can follow this same pattern.
Branching in experiments
When running experiments that compare many different approaches, it is likely you’ll need to run different procedures for different parameter sets (as different stages may be necessary, for example in training an sklearn algorithm versus a model implemented in pytorch.) Since experiment scripts are arbitrary, you can create parameters intended solely for use in the experiment itself, and simply branch based on those parameters for each given parameter set:
def run(param_sets: List[Params], mngr: ArtifactManager):
proc1 = Procedure([get_data, clean_data, train_sklearn_alg, test_model], mngr)
proc2 = Procedure([get_data, clean_data, train_pytorch, test_model], mngr)
for param_set in param_sets:
# we assume "model_type" is one of the arguments
if param_set.model_type == "sklearn":
proc1.run(param_set)
elif param_set.model_type == "pytorch":
proc2.run(param_set)
Procedure([compile_results], mngr).run(None) # an appropriately constructed
# aggregate stage can still run
# to compare outputs across multiple
# procedures.
Using the git commit hash from reports
Every time an experiment is run, the experiment store keeps track of the current git commit hash.
If you need to be able to exactly reproduce an experiment, ensure that all code is committed before
the run, and run it with the --full-store flag (see the Full stores (--store-full, --dry-cache) section.)
The output report from your run will contain the git commit hash in the top metadata fields, as well
as the command to reproduce it with the correct cache.
To re-run, checkout that hash in git, and enter the reproduce command given in the report:
git commit
experiment some_experiment -p some_params --store-full
# review the report to get the appropriate run reproduce command and the git commit hash
# to exactly reproduce:
git checkout [COMMIT_HASH]
experiment some_experiment -p some_params --cache data/runs/[RUN_REF_NAME] --dry-cache
Softlinking data directories
If dealing with very large data, in order to not have to mess with cache paths it can sometimes be useful to softlink the data/cache and data/runs. If you have one or more starting datasets that you want your experiments to have access to, you could softlink it to data/raw
In order to make these easy to work with, it’s recommended to make a shell script or makefile that runs something like:
# link_data_dirs.sh
ln -s [some_cache_path] ./data/cache
ln -s [some_runs_folder] ./data/runs
ln -s [some_datasets_path] ./data/raw