CLI Guide
The primary means of running Curifactory is through the provided experiment CLI.
The three main tasks that can be run through this command line tool are:
Run an experiment.
List available experiments and parameters.
Serve the local report HTML files via python simple web server.
Running an experiment
Running the experiment tool always checks for a local curifactory_config.json for
relevant paths, see Configuration.
Running an experiment requires the name of an experiment file (minus the .py)
and at least one parameters file (minus the .py) via the -p argument. Note that the filenames
you pass are not full paths, they are only filenames within the paths specified in the
configuration (by default an /experiments/ and /params/ folder.)
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE]
The above command calls the get_params() from the PARAMS_FILE and passes the returned list
of ExperimentParameters instances into the run() function inside EXPERIMENT_FILE.
For example, with the following project setup:
/params
__init__.py
sklearn_algs.py
/experiments
compare_models.py
We would run the experiment from the project root directory with:
experiment compare_models -p sklearn_algs
You can optionally specify multiple parameter files, where the combined returned ExperimentParameters
lists from each file are placed into a single list and passed into the experiment.
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE1] -p [PARAMS_FILE2]
Curifactory checks both the params module path as well as the experiments module
path for files with valid get_params() files, so an experiment file that
has this function may also be used in a -p flag.
If you wish to run an experiment that has a get_params() defined and you
want to use it as your parameter set, you can simply use
experiment [EXPERIMENT_FILE]
Which curifactory will automatically expand to
experiment [EXPERIMENT_FILE] -p [EXPERIMENT_FILE]
Filtering to specific parameter sets
When there are parameter files that return large lists of Params instances, it can be useful
to run only a small subset of them to update specific results or for testing purposes.
This can be done with any of the --names|-n, --indices, or --global-indices arguments.
For the following examples, assume we have a single parameters file returning three argsts:
def get_params() -> List[Params]:
return [
Params(name="baseline_knn", ...)
Params(name="baseline_svm", ...)
Params(name="baseline_mlp", ...)
]
Using the --names (or -n) argument will only run the experiment with parameter sets that have one of the
specified names. For example:
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] -n baseline_knn -n baseline_svm
will pass only the knn and svm parameter sets into the experiment file. The same thing can be achieved
with the --indices argument, specifying what indices of the parameter sets to run from each
given parameter file. (Specifying multiple parameter files and multiple indices will
run those indices from every parameter file.)
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --indices 0 --indices 1
You can also specify ranges within a single --indices argument (note that the lower
bound is inclusive and the upper bound is exclusive):
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --indices 0-2
Finally, the --global-indices specifies which indices out of the entire combined list of
parameter sets to run. This is applicable when multiple parameter files are specified, and means that
the order in which you specify them will matter. (This argument can handle ranges the same way as
indices.)
Caching controls
Caching is an important aspect in Curifactory, allowing stages to save and automatically reload data without needing to rerun portions of the code. This is also useful for sharing entire experiment runs. There are several different command line arguments for influencing how caching works.
Specifying cache directory (-c, --cache)
By default, the directory used for raw caching is set in the curifactory_config.json. For
individual experiment runs, this can be changed by providing the -c, --cache argument,
for which all cache data will be saved and loaded from the specified directory. This is particularly
relevant if attempting to reproduce somebody else’s experiment and they have a --store-full
run folder. (See the Full stores (--store-full, --dry-cache) section below .)
Overwriting cached data (--overwrite, --overwrite-stage)
Any changes made to the arguments running through an experiment will result in a different parameter hash
and thus new cached files than previous runs. However, code changes will not force a cache overwrite,
so in order to prevent inconsistent or incorrect data, you can force the experiment to ignore any
previously cached data by specifying the --overwrite argument.
If only specific stages have changed, rather than overwriting all cached data you can force a stage
run of only desired stages with the --overwrite-stage argument:
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --overwrite-stage model_train --overwrite-stage model_test
The above example will ignore cached values only for the model_train and model_test stages.
Note that overwriting a stage in the middle of an experiment will not cause later stages to also overwrite,
meaning that outdated data may still be in use.
Full stores (--store-full, --dry-cache)
Curifactory can collect all relevant data for a single experiment run and keep it in a
run-specific folder, known as a --store-full run. This folder has all cached data
from the run, a copy of the log and output report, and system environment information. This
is useful for keeping finalized versions of experiments, for distributing runs to others for
analysis, or for simply allowing easier reproduction of a specific set of results.
For experiment reproduction, or running an experiment using an existing run-specific cache,
it is also useful to use the --dry-cache argument, which allows stages to read files
from the cache but prevents them from writing to it. This also allows you to specify stage overwrites
to force specific stages to run without overwriting any of the previously cached files.
In practice, this looks something like the following:
# run the experiment and store results in a run folder
# by default, this is data/runs/[RUN_REFERENCE_NAME]/
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --store-full
# reproduce the run
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --cache data/runs/[RUN_REFERENCE_NAME] --dry-cache
Lazy cache objects (--lazy, --ignore-lazy)
As discussed in the getting started documentation, lazy cachers keep objects out
of memory as much as possible, loading them only if directly accessed. Normally,
lazy cache objects are specified by initializing a stage output name with the
Lazy class, but you can tell curifactory to assume all outputs are lazy
by running:
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --lazy
Note that lazy cache objects must have cachers specified to work. Since a stage can be specified without giving cachers, using this flag will automatically apply a pickle cacher to any outputs that do not have a specified cacher. While this should work for any pickleable objects, it may fail if any stages output something that can’t be correctly pickled.
Similarly, if running stages that have lazy objects but in an environment where
keeping them in memory isn’t a big deal, you can turn off all lazy caching with
the --ignore-lazy flag. In cases when dealing with large objects that
are expensive to repeatedly save and reload, this can potentially speed up an experiment
run:
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --ignore-lazy
Parallel runs
In situations where you may have a large number of argsets to run through a lengthy experiment, Curifactory can run many instances of the experiment in parallel, using the multiprocessing library.
Running an experiment with the --parallel 4
argument will divide up the entire list of parameter sets into four ranges of global argument indices,
spawn four processes, and run the experiment in each, passing in the range for that process. After
all processes complete, the experiment is run again with all parameter sets. The idea is that, assuming
caching is done in every important stage, all relevant data for each stage in the full run has
already been cached from the individual runs done via multiprocessing, and so only final aggregate
stages need to be re-run against the full set of records.
This approach is very loosely equivalent to the below commands, assuming the parameters file returns 8 argsets:
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --parallel 4
# the above translates* into running the following in separate processes:
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --parallel-mode --global-indices 0-2
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --parallel-mode --global-indices 2-4
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --parallel-mode --global-indices 4-6
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --parallel-mode --global-indices 6-8
# with a final full run to handle the report and any aggregate stages:
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE]
Note that running the above commands is not actually equivalent to running the experiment
with --parallel. Using multiprocessing creates lock and queue variables that are used
to ensure the processes aren’t stepping on each other, which can occur if you attempt to manually
run these in parallel using the --parallel-mode flag.
In order for parallel runs to be effective, there are a few assumptions about the experiments:
There are few/low-compute-intensity aggregate stages towards the end of the experiment. Usually these are only done for final comparison/result analysis steps.
All compute-heavy stages cache their outputs. The intent for this approach is that the final full run doesn’t need to run any compute.
The type of parallelization you’re going for is on the very coarse experiment/argset level. This does not split computation at the stages or provide any sort of parallelization beyond running multiple argsets through the same experiment at the same time.
Listing experiments and parameters
You can get a list of valid experiment files and parameter files in the commandline by running:
experiment ls
This will check every file in the experiments folder for files containing a run() function
and every file in the parameters folder for files containing a get_params() function. Note
that Curifactory attempts to import all files in order to check for potential errors on import, so
standard warnings for if you’re importing somebody else’s code applies. This command can also take
a while to run if any of the files have a large number of slow imports.
The output listing looks something like:
EXPERIMENTS:
example_experiment - Some experiment description
example_experiment2
PARAMS:
example_params - Basic set of parameters
example_params2 - Fancier parameters
The descriptions after the - for each entry in the listing are directly parsed from any docstrings at the top of the relevant files, this is particularly useful when there are a large number of experiment and/or parameter files.
Experiment run notes
You can provide notes for an experiment run with the --notes flag. The
idea for these is to be vaguely like git commit messages, in that if the notes
span multiple lines, the first line will be the shortform version (displayed on
the report index page) and the remainder of the lines will render in full on the
experiment preport itself.
You can either specify notes inline:
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --notes "This is a note for this experiment run"
Or simply specifying the notes flag by itself will open a text editor for you to enter the notes.
experiment [EXPERIMENT_FILE] -p [PARAMS_FILE] --notes
Once the text editor is saved and exited, the note content will
be used for the run notes. If a EDITOR environment variable is set,
curifactory will attempt to open that editor, otherwise it will run through a
list of editors and open the first one it finds.
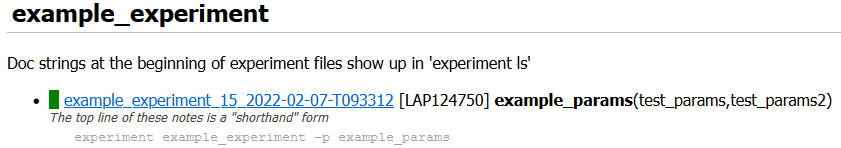
The first line of the notes (if multi-line) shows up with the run in the report index in italics.

The entire notes content shows up in a “Notes” section under the info block in the run report.
Hosting HTML reports
Every experiment that runs to completion generates an HTML run report in the reports/
folder, and updates the top level reports index.html. These files can be served with:
experiment reports
By default, this serves them on port 8080, but this can be configured with the --port flag:
experiment reports --port 6789
The IP the server will accept connections from can also be configured with the
--host flag. By default this is 127.0.0.1, only allowing localhost
connections.
experiment reports --host 0.0.0.0
Note that if you run experiments on multiple machines and transfer all of the
reports to the same folder, the report index will not accurately reflect them.
You can use experiment reports --update to regenerate this index based
on all discovered folders in your reports directory.
Full reference
Below is the full set of flags that can be used with the experiment command.
Parameter names (-p, --params`)
The name of a python parameters file with a get_params() function. You can specify multiple
parameter files with multiple -p flags, which will combine all returned parameter sets from
all parameter files into a single parameter set list.
Note that you do not include the actual path or .py, the string is directly used in the import as a
submodule of the params module/folder.
Example:
experiment some_experiment_name -p my_params -p my_other_params
Parameter set names (-n, --names)
Run the experiment with only argsets with the specified names (the name defined in the ExperimentArgs
instance) given with this flag. Use only one name per flag, but as with the parameter names, this
flag can be specified multiple times. See Filtering to specific parameter sets above.
Example:
experiment some_experiment_name -p my_params -n base_knn -n base_randomforest
Argument set indices (--indices)
Run the experiment with only argsets with the specified indices given with this flag within
each specified parameters file. Indices can either be specified as individual numbers or as
ranges in the format [inclusive lower index]-[exclusive upper index]. As with
--names, this flag can be specified multiple times. See Filtering to specific parameter sets above.
Example:
experiment some_experiment_name -p my_params --indices 0-2 --indices 2
Global arument set indices (--global-indices)
Run the experiment with only argsets with the specified indices given with this flag out of
the total list of args from all specified parameters files. Indices can either be specified
as individual numbers or as ranges in the format [inclusive lower index]-[exclusive
upper index]. As with --names, this flag can be specified multiple times. See
Filtering to specific parameter sets above.
Example:
experiment some_experiment_name -p my_params --global-indices 0-2 --global-indices 2
Parallel (--parallel)
Runs the experiment in parallel across the specified number of processes. This automatically divides up the arguments into a roughly even number per process, so each process only runs the experiment with those argsets. After each process completes, the full experiment is run again, but ideally all necessary data is already cached. See Parallel runs.
Example:
experiment some_experiment_name -p my_params --parallel 2
Parallel mode (--parallel-safe)
Suppresses writing to the experiment store and output report while still running and caching results. This could in theory be used for writing your own paralleliztion, but be warned that the actual internal parallelization handles file locks appropriately, while this flag does not. See Parallel runs.
Suppress logging (--no-log)
Specifying this flag will disable writing a console log file to the logs/ directory.
Overwrite cache (--overwrite)
Specifying this flag ignores any existing cached data and will force all computation to run, overwriting all data. See Overwriting cached data (--overwrite, --overwrite-stage).
Overwrite cache for stage (--overwrite-stage)
Ignore only the cached outputs for the specified stages. You can specify this flag multiple times to ignore the cache for several stages. Note that any later computations are not automatically also overwritten, so take care with nondeterministic outputs in the middle of an experiment. See Overwriting cached data (--overwrite, --overwrite-stage).
Example:
experiment some_experiment_name -p my_params --overwrite-stage train_model --overwrite-stage test_model
Full store (-s, --store-full)
Keep a full copy of all cached data, environment information, log, and output report from an
experiment run in a run-specific folder, determined by the run_path in the configuration.
See Full stores (--store-full, --dry-cache).
Cache directory (-c, --cache)
Specify what directory to use for reading and writing cached data, if it differs from the configuration value. This is useful if using somebody else’s cached run. See Caching controls.
Example:
experiment some_experiment_name -p my_params --cache data/runs/some_specific_run_foldre --dry-cache
Force lazy caching (--lazy)
Treat all stage outputs as Lazy objects. Pickle cachers will be
injected for any outputs that have no cacher specified. See Caching controls.
Force no lazy caching (--ignore-lazy)
Treat lazy outputs like regular outputs, keeping them in memory instead. See Caching controls.
Include debug logs (-v, --verbose)
Include debug messages in the output logs if specified.
Progress bars (--progress)
Output fancy rich progress bars for every record and overall experiment execution. Note that if you have code using TQDM progress bars, this can sometimes conflict and cause weird formatting issues, which is why it’s not enabled by default.
Only print experiment map (--map)
Specifying this flag only runs the pre-mapping phase of an experiment and then exits, printing out a summary of all records, stages, and artifacts with their respective cache statuses. This is useful for determining (before actually running) whether an experiment has everything cached or not.
Note that if you run this with --verbose, it will also print out the exact set of
stages it plans to execute in DAG model.
Run experiment purely linearly instead with DAG (--no-dag)
To disable the DAG-based execution of an experiment, specify -no-dag. When
running an experiment normally (in DAG mode), there is an experiment pre-mapping
step that steps through your entire experiment and executes only the decorator portion
of each stage, in order to map out what artifacts are inputs/outputs, which ones
are already cached, and what stages actually need to execute in order to produce
any final leaf artifacts. In some cases either with weird stage setups, or non-stage
functions that take a long time to run or have side-effects, use --no-dag to
eliminate this pre-mapping step and simply execute all the stages linearly (short-circuiting
only based on cache status rather than map status.)
Note that progress bars from --progress won’t work in --no-dag mode.
Suppress console log output (--quiet)
Make the output a little less busy and don’t include logging messages. Note that
this will still render the progress bars, unless you also specify
--no-dag.
Suppress terminal colors in case of unrecognized non-support (--no-color)
Not all terminals support color well, and the output is full of fun colors from Rich. Suppress them with this flag.
Suppress rich formatting of logging (--plain)
Turn off using Rich logging formatter. The logs output to the terminal will look exactly as they’re written into the log files.
Suppress all file output (--dry)
Runs the experiment without outputting or altering any files, including logs, experiment store, parameter registry, and cached data.
Suppress writing to cache (--dry-cache)
Allows reading from the cache but will not write any cache files. This is useful in combination with a non-default cache directory if reading somebody else’s cached run. See Caching controls.
Include errors in log files (--log-errors)
Specifying this will record any errors and stack traces in the output log files by redirectring STDERR. Note that some libraries use STDERR for non-error messages, such as TQDM’s progress bars. (In some cases this may output a lot of extra lines into the log.)
Custom cache name prefix (-n, --name)
Give the cached values an explicit prefix as opposed to the experiment name.
Example:
experiment some_experiment_name -p my_params --name final_run
The above will output cache filenames starting in final_run_[ARGHASH]... rather than
some_experiment_name_[ARGHASH]. This can be useful for tracking specific runs without
using --full-store.
Experiment run notes (--notes)
Include notes in the run report for an experiment run.
Example:
experiment some_experiment_name -p my_params --notes "A simple test run."
Alternatively use a system text editor to enter notes:
experiment some_experiment_name -p my_params --notes
See Experiment run notes.
Export experiment in docker container (--docker)
After the experiment completes, build a docker image with a complete copy of the current
codebase, all data from the experiment run, and the output notebook. Assuming the default
dockerfile that is created when running curifactory init, the resulting image, when
run, hosts a file server with the data cache as well as the run notebook.
Note that a wheel of Curifactory will need to be built, placed in the docker folder and referenced in the dockerfile in order for the notebook to run correctly. (This will be addressed in a later version.)
Export experiment explorer as a jupyter notebook (--notebook)
After the experiment completes, write a jupyter notebook with information about the run and basic template code to load and explore the cached data.
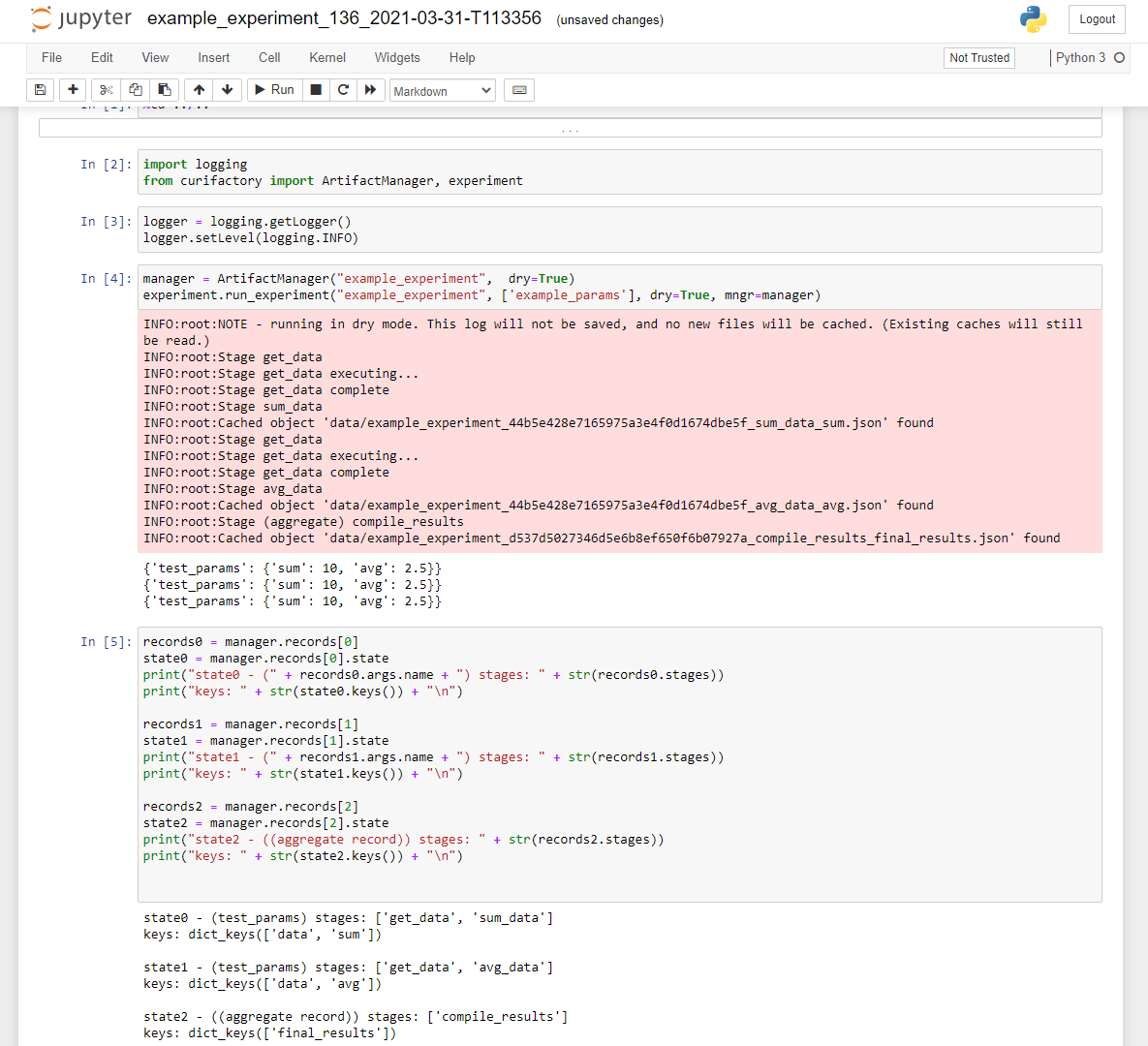
An example output notebook generated after running the experiment.
This works by writing the code to re-run the experiment with the same parameters and experiment name, meaning all relevant data should already be cached. This results in a set of records in memory that should mirror the previous run, allowing live exploration of the states.
Note that this requires an environment with ipynb-py-convert accessible from the command line,
which should come with any jupyter installation.
Serve HTML reports on a specified port and host (--port, --host)
These flags only applies to the experiment reports command, specifying which port to
serve the HTML reports on and what addresses to accept connectiosn from. See
Hosting HTML reports.